Ortega A, Gonzalo-Asensio J, García-Del Portillo F.
 Small non-coding regulatory RNAs (sRNAs) have been studied in many
bacterial pathogens during infection. However, few studies have focused
on how intracellular pathogens modulate sRNA expression inside
eukaryotic cells.Here, we monitored expression of all known sRNAs of Salmonella enterica serovar Typhimurium (S. Typhimurium) in bacteria located inside fibroblasts, a host cell type in which this pathogen restrains growth. sRNA sequences known in S. Typhimurium and Escherichia coli were searched in the genome of S. Typhimurium
virulent strain SL1344, the subject of this study. Expression of 84
distinct sRNAs was compared in extra- and intracellular bacteria.
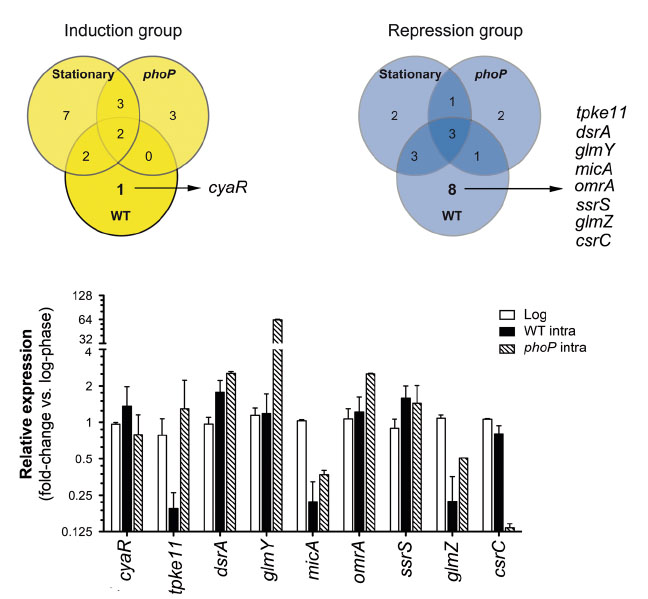
Non-proliferating intracellular bacteria upregulated six sRNAs,
including IsrA, IsrG, IstR-2, RyhB-1, RyhB-2 and RseX while repressed
the expression of the sRNAs DsrA, GlmZ, IsrH-1, IsrI, SraL, SroC,
SsrS(6S) and RydC. Interestingly, IsrH-1 was previously reported as an
sRNA induced by S. Typhimurium inside macrophages. Kinetic
analyses unraveled changing expression patterns for some sRNAs along the
infection. InvR and T44 expression dropped after an initial induction
phase while IstR-2 was induced exclusively at late infection times (>
6 h). Studies focused on the Salmonella-specific sRNA RyhB-2 revealed
that intracellular bacteria use this sRNA to regulate negatively YeaQ, a
cis-encoded protein of unknown function. RyhB-2, together with RyhB-1,
contributes to attenuate intracellular bacterial growth. To our
knowledge, these data represent the first comprehensive study of S. Typhimurium
sRNA expression in intracellular bacteria and provide the first
insights into sRNAs that may direct pathogen adaptation to a
non-proliferative state inside the host cell.
Small non-coding regulatory RNAs (sRNAs) have been studied in many
bacterial pathogens during infection. However, few studies have focused
on how intracellular pathogens modulate sRNA expression inside
eukaryotic cells.Here, we monitored expression of all known sRNAs of Salmonella enterica serovar Typhimurium (S. Typhimurium) in bacteria located inside fibroblasts, a host cell type in which this pathogen restrains growth. sRNA sequences known in S. Typhimurium and Escherichia coli were searched in the genome of S. Typhimurium
virulent strain SL1344, the subject of this study. Expression of 84
distinct sRNAs was compared in extra- and intracellular bacteria.
Non-proliferating intracellular bacteria upregulated six sRNAs,
including IsrA, IsrG, IstR-2, RyhB-1, RyhB-2 and RseX while repressed
the expression of the sRNAs DsrA, GlmZ, IsrH-1, IsrI, SraL, SroC,
SsrS(6S) and RydC. Interestingly, IsrH-1 was previously reported as an
sRNA induced by S. Typhimurium inside macrophages. Kinetic
analyses unraveled changing expression patterns for some sRNAs along the
infection. InvR and T44 expression dropped after an initial induction
phase while IstR-2 was induced exclusively at late infection times (>
6 h). Studies focused on the Salmonella-specific sRNA RyhB-2 revealed
that intracellular bacteria use this sRNA to regulate negatively YeaQ, a
cis-encoded protein of unknown function. RyhB-2, together with RyhB-1,
contributes to attenuate intracellular bacterial growth. To our
knowledge, these data represent the first comprehensive study of S. Typhimurium
sRNA expression in intracellular bacteria and provide the first
insights into sRNAs that may direct pathogen adaptation to a
non-proliferative state inside the host cell.
